 |
 |
(4.18) |
 |
![$\displaystyle = \mathbf{B}\boldsymbol{\phi}[ \mathbf{h}(t) ] + \mathbf{C}\mathbf{s}(t) + \mathbf{b} + \mathbf{n}_x(t) \, ,$](img273.png) |
(4.19) |
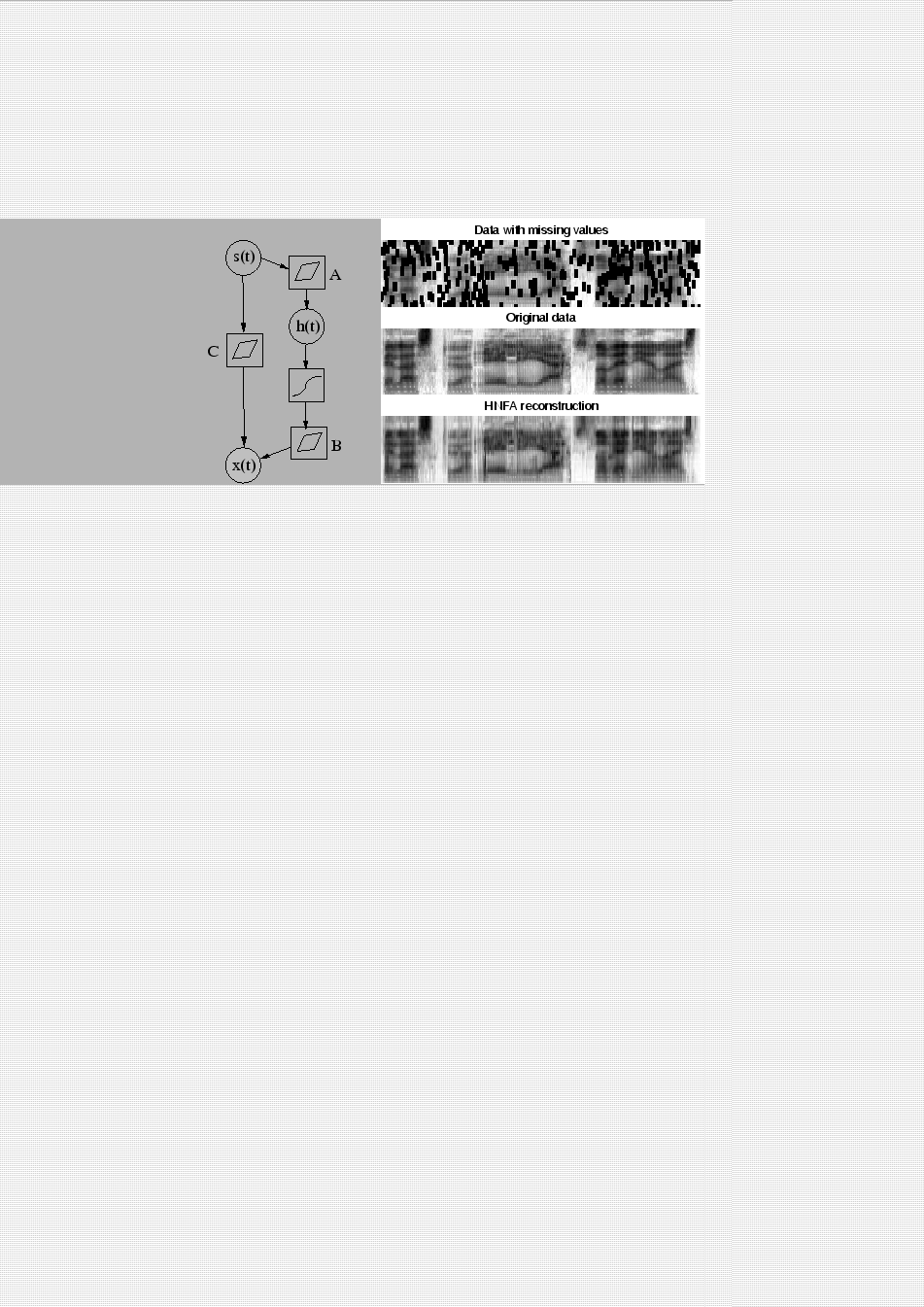
In hierarchical nonlinear factor analysis (HNFA) (Raiko, 2001; Valpola et al., 2003b), there are a number of layers of Gaussian variables, the bottom-most layer corresponding to the data. There is a linear mixture mapping from each layer to all the layers below it. The middle layer variables are immediately followed by a nonlinearity. The model structure for a three-layer network using Bayes Blocks is depicted in the left subfigure of Figure 4.6. Model equations are
 and
and
 are Gaussian noise terms and the
nonlinearity
are Gaussian noise terms and the
nonlinearity
 operates on each element
of its argument vector separately. This activation function has the universal approximation property as well (see Stinchcombe and White, 1989, for proof).
Note that the short-cut mapping
operates on each element
of its argument vector separately. This activation function has the universal approximation property as well (see Stinchcombe and White, 1989, for proof).
Note that the short-cut mapping  from sources
to observations means that hidden nodes only need to model the
deviations from linearity.
from sources
to observations means that hidden nodes only need to model the
deviations from linearity.
 |
HNFA has latent variables
 in the middle layer, whereas in
nonlinear FA, the middle layer is purely computational. This results
in some differences. Firstly, the cost function
in the middle layer, whereas in
nonlinear FA, the middle layer is purely computational. This results
in some differences. Firstly, the cost function
 in HNFA is
evaluated without resorting to approximation, since the required
integrals can be solved analytically. Secondly, the computational
complexity of HNFA is linear with respect to the number of sources,
whereas the computational complexity of nonlinear FA is quadratic.
HNFA is thus applicable to larger problems, and it is feasible to use
even more layers than three. Also, the efficient pruning facilities
of Bayes Blocks allow determining whether the nonlinearity is
really needed and pruning it out when the mixing is linear, as
demonstrated by Honkela et al. (2005).
in HNFA is
evaluated without resorting to approximation, since the required
integrals can be solved analytically. Secondly, the computational
complexity of HNFA is linear with respect to the number of sources,
whereas the computational complexity of nonlinear FA is quadratic.
HNFA is thus applicable to larger problems, and it is feasible to use
even more layers than three. Also, the efficient pruning facilities
of Bayes Blocks allow determining whether the nonlinearity is
really needed and pruning it out when the mixing is linear, as
demonstrated by Honkela et al. (2005).
The good properties of HNFA come with a cost. The simplifying
assumption of diagonal covariance of the posterior approximation,
made both in nonlinear FA and HNFA, is much stronger in HNFA
because it applies also in the middle layer variables
 .
Publication II compares the two methods in
reconstructing missing values in speech spectrograms. As seen in the
right subfigure of Figure 4.6, HNFA is able to
reconstruct the spectrogram reasonably well, but quantitative comparison
reveals that the models learned in HNFA are more linear (and thus in
some cases worse) compared to the ones learned in nonlinear FA.
.
Publication II compares the two methods in
reconstructing missing values in speech spectrograms. As seen in the
right subfigure of Figure 4.6, HNFA is able to
reconstruct the spectrogram reasonably well, but quantitative comparison
reveals that the models learned in HNFA are more linear (and thus in
some cases worse) compared to the ones learned in nonlinear FA.