 |
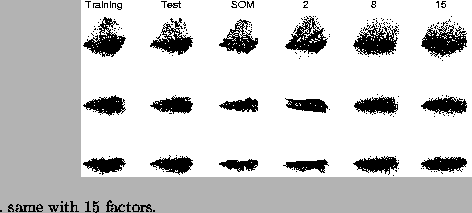
Figures 5 and 7 show some projections of the data and the models. The SOM manifold looks quite curly. NFA manifold with two factors is also rolled up, but in a smoother way. Important differences between high dimensional NFA manifolds are not visible in these projections.
 |
As expected, the SOM performs the best for clustered data and NFA performed better than FA. Results show that NFA is closer to FA than to the SOM. The greatest problem of the SOM is that the number of parameters scales exponentially with the number of intrinsic dimensions of the data manifold, which leads to bad generalisation. The NFA model does not work well, if the data is clustered. It is hard to find the function that shapes a Gaussian continuum to clusters, but when the data forms a continuum, too, the NFA model is more appropriate.
The NFA algorithm searches only for local optima, so multiple runs with different initialisations would have better chances globally. The stability should be guaranteed and some speedups could be made for the algorithm to be a ready-to-use tool.
Even though the learning algorithm requires a lot of processing, the NFA model, when it has learned, can be applied in real time. The computational complexity of the algorithm scales to the product of number of factors and hidden neurons per batch iteration. By contrast, the number of map units in a SOM scales exponentially as a function of the dimensionality of the map. NFA is best suited for fairly strongly nonlinear problems with an intrinsic dimension of the order of ten.
When compared to the SOM, factor analysis models are simpler in terms of the number of parameters. While even the largest NFA model had about 3000 parameters, the SOM had about 50000 parameters. This can be seen in Figure 5 from the fact that NFA did not capture all the finest details in the data set. Experimental results support the statement that small number of parameters enhances the ability to generalise.